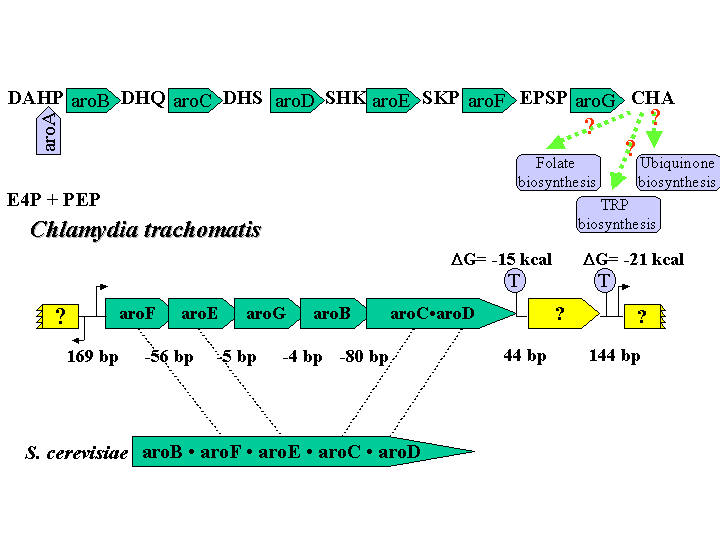
Examination of the aromatic aminoacid biosynthesis pathway in Chlamydia trachomatis revealed that the chorismate biosynthesis portion of the pathway was intact, however most enzymes necessary for the conversion of chorsimate to the 3 branch aromatic aminoacids were not present (only tyrB, trpA/B and trpF have been identified in Chlamydia trachomatis). This truncated aromatic amino acid biosynthesis pathways in Chlamydia trachomatis give rise to end product -chorismate that are used as intermediates for other biosynthetic pathways. So the ubiquinone, folate, menaquinone and enterobactin biosynthesis pathway have been searched in Chlamydia trachomatis. About half of the ubiquinone and folate biosynthesis pathways genes appear in Chlamydia trachomatis, however almost all of genes for menaquinone and enterobactin biosynthesis are missing. In folic acid biosyntheses pathway, Chlamydia trachomatis is missing pabA, pabB (encoding aminodeoxychorismate synthase) and pabC (encoding aminodeoxychorismate lyase), which catalyze the reaction from chorismate to p-aminobenzoate. We suppose this means that Chlamydia trachomatis might depend on the host to supply PABA for folic acid biosynthesis. In ubiquinone biosynthesis pathway, Chlamydiae trachomatis contain homologs of ubiA (encoding 4-hydroxybenzoate octaprenyltransferase); ubiX (encoding 3-octaprenyl-4-hydroxybenzoate decarboxylase) and ubiE (2-octaprenyl-6-methoxy-1, 4-benzoquinone methylase), the genes encoding the rest 5 steps of reactions are missing.
It is very interesting that the chorismate biosynthetic pathway genes in CT are in the same operon except the first gene aroA is at difference place. In many microbial eukaryotes, the aroM gene from fungi encodes a single polypeptide that catalyses five consecutive steps of the chorismate biosynthetic pathway. If the genes encoding enzymes in the common-pathway portion of aromatic biosynthesis are named in order of reaction sequence (i.e., aroA-aroG), the domain order in aroM gene is aroB*aroF*aroE*aroC*aroD.

The compact intergenic spacing from aroF to aroC*D and the translationally coupled (5 genes) suggest the possibility of a single transcriptional unit. Although these genes order in fungi is different from in CT, aroF-aroE and aroC-aroD are often conventionally organized. These observations strongly suggest that the genes are related to common ancestors and that the genes encoding the pentafunctional proteins evolved by multiple gene fusions.