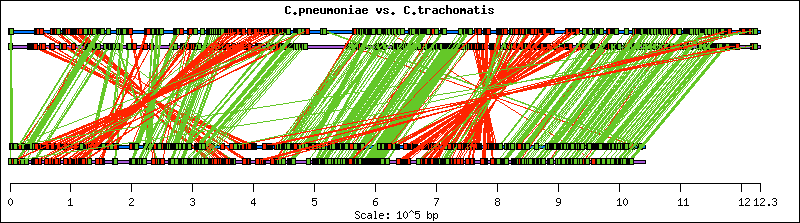

A comparison of the relative locations of conserved orthologous regions between the bacterial genomes of C.pneumoniae and C.trachomatis was performed using the BLAST2 database comparison tool. Significance was determined using an E-value threshold of 0.001. A region was considered conserved if at least two consecutive genes in one genome were orthologous to at least two genes that were consecutive in the other genome.
This visual genome comparison reveals complex rearrangements between C.trachomatis and C.pneumoniae, suggesting inversions located at the genes forming crossed red lines. An inversion, defined by Lewin (Genes V, 1994) as "a chromosomal change in which a segment has been rotated by 180 degrees relative to the regions on either side and reinserted," creates red lines that cross to form an "X" pattern. Green parallel lines result from no change in gene order and direction between the two genomes. Differences in the angles of parallel regions may suggest insertions or deletions in one genome, or may indicate numerous duplications in one genome.
Kalman et al. (1999) also observed a high level of genetic rearrangement between C.trachomatis and C.pneumoniae. The large proportion of orthologs revealed functional conservation between the two genomes, and, although rearrangement had occurred, the order of many regions was conserved. This suggests that certain regions require gene proximity in order to be expressed appropriately, but that overall region order did not alter the function of these conserved genes. Genome comparisons performed upon other bacterium have also revealed the wide-spread characteristic of complex rearrangements coupled with conserved function (Watanabe et al., 1996, Siefert et al., 1997, Himmelreich et al., 1997, Casjens, Annu Rev Gen, 32:339-77, 1998).